|
Abstract
The methods of linear algebra are used to distinguish between different gaits.
A probability space is a triplet with:
a sample space of all possible outcomes;
a set of events that is a set of subsets of ;
a probability function for each event.
Rather improperly named, a random variable , is a function defined on a sample space with values in a measurable space (e.g., ). For some measurable subset , the probability of is
A stochastic process is indexed collection of random variables. Often the index is time, and . A centered stochastic process has mean value zero
with the expectation operator.
The Karhunen-Loève theorem affirms the existence of a canonical description of a stochastic process as a linear combination of random variables with time-dependent coefficients , or, conversely, as a linear combination of time-varying functions with random coefficients
As will be explained in detail in the STC module, the singular value decomposition is a discrete form of the Karhunen-Loève theorem. Let the matrix denote multiple samples of some real-valued, centered stochastic process
There exist orthogonal matrices , , and the quasi-diagonal positive matrix such that
known as the singular value decomposition (SVD). Introducing the column vectors of
the SVD can be rewritten in two important forms:
The covariance of two centered random variables is
typically approximated through a statistic from observations
The covariance matrix of centered random variables has entries
and is expressed as the matrix product
By construction the covariance matrix is symmetric positive definite (spd) and therefore admits an orthogonal eigendecomposition
Assume (by re-labeling if necessary) that . It is often the case that eigenvalues dominate over all others (the strongly correlated modes)
and the correlation matrix is approximated by the first rank-1 updates
The dominant correlated modes form a natural basis set for analysis of data. Rather than solving the covariance matrix eigenproblem the SVD of is used since
Consider the data obtained from many individual gait measurements (either from different individuals or at different times for the same individual). The goal is to identify differences w.r.t. a mean gait and use those differences to (1) identify either an individual or (2) a particular type of walking (climbing stairs versus level walking). The procedure is demonstrated here for the second problem.
octave>
dir='/home/student/courses/MATH590/NUMdata';
chdir(dir);
LastName='Mitran';
d=textread(strcat(LastName,'.data'));
mu=max(size(d))/7; disp(mu)
octave>
data=reshape(d,[7,mu])';
data(1:6,1:7)
octave>
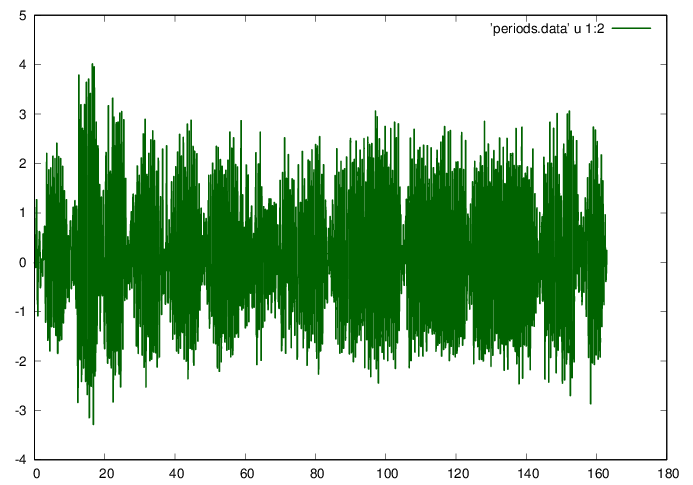
Interpolate to obtain even-spaced data, and plot the data.
octave>
t0=data(1,1); t1=data(mu,1); ni = 2^ceil(log2(mu));
dt=(t1-t0)/ni;
ti=(0:ni-1)*dt; ti=ti';
ai=interp1(data(:,1),data(:,3),ti);
fid=fopen('periods.data','w');
fprintf(fid,'%f %f\n',[ti ai]');
fclose(fid);
octave>
GNUplot]
cd '/home/student/courses/MATH590/NUMdata'
set terminal postscript eps enhanced color
set style line 1 lt 2 lc rgb "0x00006400" lw 3
plot 'periods.data' u 1:2 w l ls 1

GNUplot]
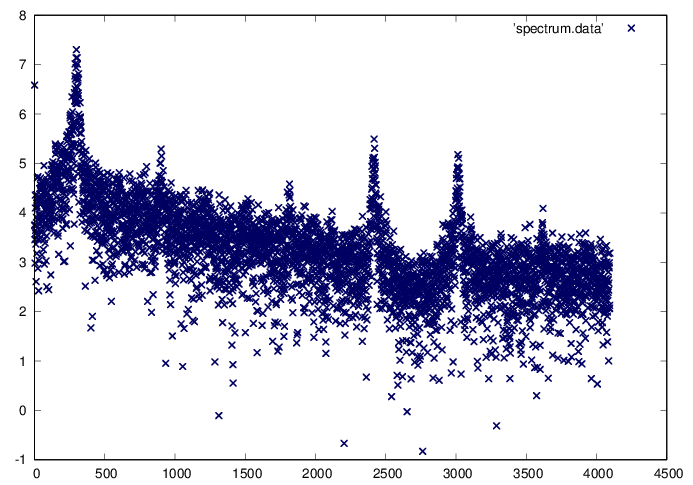
Find the Fourier spectrum of the vertical acceleration data.
octave>
Ai=fft(ai); PAi=log10(Ai.*conj(Ai));
fid=fopen('spectrum.data','w');
fprintf(fid,'%f\n',PAi(1:ni/4));
fclose(fid);
octave>
GNUplot]
cd '/home/student/courses/MATH590/NUMdata'
set terminal postscript eps enhanced color
set style line 1 lt 2 lc rgb "0x00000064" lw 3
plot 'spectrum.data' ls 1
GNUplot]
There are several peaks within the Fourier spectrum. Seek the peak corresponding to a natural step period of approximately . This turns out to be close to the global peak as shown in the following calculations
octave>
ks=floor((t1-t0)/(0.5))
octave>
[PAimx imx]=max(PAi);
octave>
[PAimx imx]
octave>
N=imx-1;
octave>
Ts=(t1-t0)/N
octave>
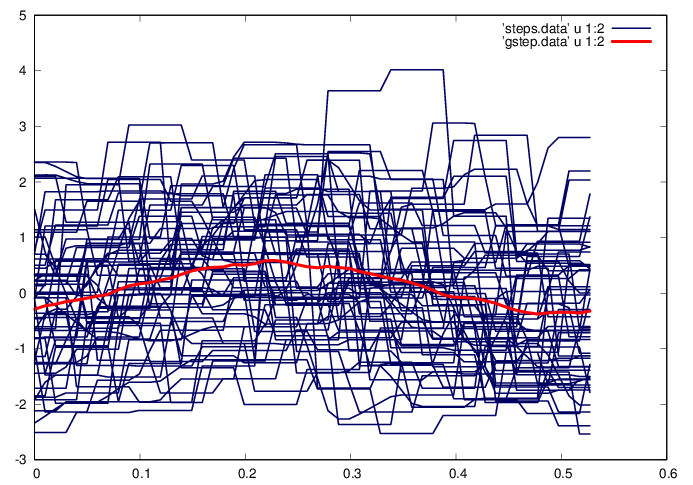
Isolate the steps, find an average step waveform , and compute the centered data matrix
octave>
m = floor(ni/N); t=(0:m-1)'*dt;
a = reshape(ai(2:m*N+1),[m,N]); g = mean(a')';
X = a - repmat(g,1,N);
octave>
fid=fopen('steps.data','w');
i=1; while(i<N)
fprintf(fid,'%f %f\n',[t a(:,i)]');
fprintf(fid,'\n');
i=i+5;
endwhile;
fclose(fid);
octave>
fid=fopen('gstep.data','w');
fprintf(fid,'%f %f\n',[t g]');
fclose(fid);
octave>
GNUplot]
cd '/home/student/courses/MATH590/NUMdata'
set terminal postscript eps enhanced color
set style line 1 lt 2 lc rgb "0x00000064" lw 3
set style line 2 lt 2 lc rgb "0x00ff0000" lw 6
plot 'steps.data' u 1:2 w l ls 1, 'gstep.data' u 1:2 w l
ls 2
GNUplot]
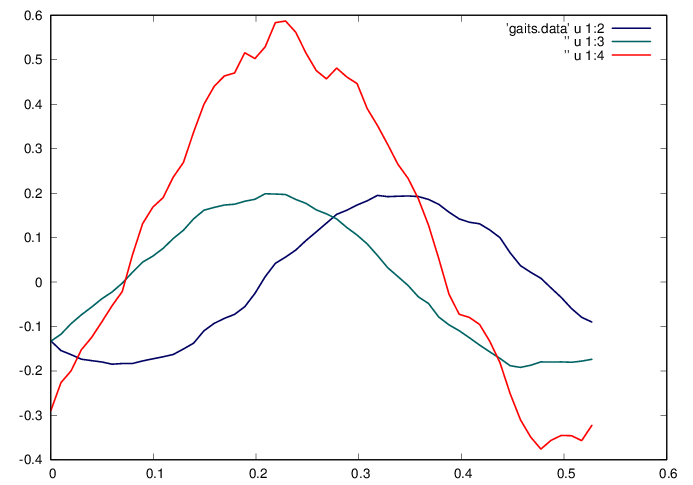
Find the natural basis for the problem by computing the SVD of , and investigate the dominant singular values
octave>
[U,S,V]=svd(X,0);
octave>
(diag(S)(1:6))'
octave>
fid=fopen('gaits.data','w');
fprintf(fid,'%f %f %f %f\n',[t U(:,1:2) g]');
fclose(fid);
octave>
From the above each step can be characterized by the first two components . Plot these modes and compare to the average gait .
GNUplot]
cd '/home/student/courses/MATH590/NUMdata'
set terminal postscript eps enhanced color
set style line 1 lt 2 lc rgb "0x00000064" lw 3
set style line 2 lt 2 lc rgb "0x00006464" lw 3
set style line 3 lt 2 lc rgb "0x00FF0000" lw 3
plot 'gaits.data' u 1:2 w l ls 1, ” u 1:3 w l ls 2,
” u 1:4 w l ls 3
GNUplot]
Find the coordinates of each step along these directions.
octave>
c=X'*U(:,1:2);
fid=fopen('coefs.data','w');
fprintf(fid,'%f %f\n',c');
fclose(fid);
octave>
size(c)
octave>
GNUplot]
cd '/home/student/courses/MATH590/NUMdata'
set terminal postscript eps enhanced color
set style line 1 lt 2 lc rgb "0x00000064" lw 3
plot 'coefs.data' u 1:2 w p ls 1
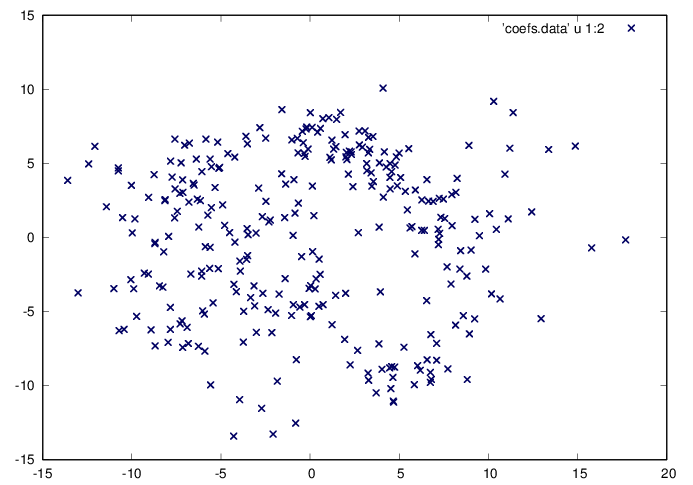
GNUplot]
At this point, an economical representation of each of the steps has been obtained, and the next question is to classify the observed results. This forms the objective of clustering analysis that relies on the mathematical theory of sets, as presented in the next module.